SATURDAY, 7 MAY 2011
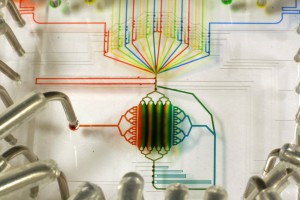
Imagine a chemistry lab the size of a postage stamp. Or a medical device that fits in your wallet with the promise of an instant health check. Perhaps you would like to scan the atmosphere for pathogens and analyse chemicals on Mars, Star Trek style? Thanks to the myriad of rapidly developing technologies that rely on microfluidics, all this is becoming possible. So, how does microfluidics work?Microfluidics is defined as the manipulation of systems in which tiny amounts of fluid flow through very narrow channels. Typically, the channels are as narrow as a human hair, less than 1 millimetre wide. The amount of fluid flowing through a device made with these narrow channels can be as little as several ‘attolitres’, which is 1/1000000000000000000th of a litre, smaller than the volume of the smallest viruses. Microfluidic devices are usually built systematically from a series of basic components, including an entrance for reagents and samples, some means of moving and mixing the fluids, and other components for producing output, such as detection or purification devices. First used as an analytical tool in chemistry, the relatively new technology offers many advantages over larger systems. Devices require a very small volume of reagents, a small amount of sample material, but are capable of delivering high resolution results at low cost and short analysis times.
Yet simply scaling down a larger system into micrometre size would not yield a working microfluidic device. This is because the behaviour of fluids at a shrunken scale is very different to that in a larger device. In the narrow channels, fluid flow is turbulent free, so there is very little mixing. If you put coffee and milk down the same channel in your microfluidic device, instead of well-mixed white coffee towards the end of the channel, you will have black coffee and milk, still separated, and flowing side by side. Only when the coffee and milk exit the channel into a big coffee cup would they resume turbulent flow, producing your milky coffee in a few seconds.
The smooth flow of fluids in microchannels means that whilst it is easy to keep two different chemicals separate even when they are flowing side by side, it is also very hard to get them to mix. Stirring is not an option because the channels are too narrow. Designers of microfluidic devices have to find ingenious ways to introduce turbulent flow and force mixture, such as building sharp bends into the channels, or using microvalves or micropumps.
The potential of microfluidics to facilitate the work of research scientists and medical professionals has led to rapid and exciting developments in the field.
Personalised medicine, rapid disease identification, forensic evidence from tiny sample; these technologies may seem decades away from realisation, but the development of lab-on-a-chip devices has helped to bring them closer to reality.
These applications depend on identifying genetic material, in particular the exact sequence that builds up a DNA strand. For example, to identify a bacterial cell unambiguously, you need to make a comparison between the DNA sequence of that cell and the sequence of a known bacterium. In many cases, the amount of genetic material available is small. In order to carry out tests, the DNA extracted from the cell of interest needs to be ‘amplified’ by a process called polymerase chain reaction (PCR).
In PCR, the original DNA sample passes through three specific temperature stages for a large number of cycles. First, a high temperature stage breaks apart the double helix of DNA in a ‘melting’ process, yielding two single-strand molecules. The temperature is lowered in the next stage, where the building blocks of DNA adhere to the single chains in a sequence-specific manner. Finally, the temperature is kept low while the building blocks are linked into a strand of DNA by an enzyme, yielding two copies of the original double helix. Through repeating many such cycles, this doubling process can exponentially amplify the sample mass while conserving the sequence of the DNA being investigated.
Traditional methods of PCR require large machines and large quantities of reagents, making this process expensive and time consuming. In addition, if the amount of sample is tiny, such is often the case in forensic investigation, then even after PCR the material is still insufficient for analysis. Errors can also be introduced by amplifying contaminants, damaging admissibility of the DNA evidence in a court of law.
PCR can be speeded up using microfluidic principles, by performing each stage of the process on one of three layers of a single chip. Fluid flow is carefully controlled in the device, allowing mixture only at desired places. The proximity of the layers also means reaction products are transferred in a fraction of the time.
Since typical lab-on-a-chip devices only require several billionths of a litre of reagents and samples, they have increased sensitivity, accurately amplifying and identifying DNA sequences where larger systems fail. As many channels and detection chambers can sit on the same chip, analyses can be done in parallel, as opposed to the linear workflow used in larger DNA analysis machines. Thus microfluidic DNA sequencing is much faster.
Microfluidic PCR has obvious implications for human genome sequencing. In the human genome project, the whole DNA sequences of several individuals were ‘read’. The traditional methods available meant the process took many years and several hundred thousand US dollars. With the ability to sequence DNA on a microfluidic chip, the process should be faster and cheaper.
Beyond looking at DNA in cells, microfluidic principles can be applied to the detection of cells and chemicals in body fluids. An example is the i-Stat device, allowing doctors to carry out bedside blood tests with almost instantaneous results using a handheld device. Only a few drops of blood are needed to carry out a series of tests. The device allows many patients to be seen and tested at the same time, as their blood is collected onto relatively cheap, single-use microfluidic cassettes. This technology is already in use in some parts of the UK; NHS workers in Kent have reported that it is especially useful for home visits.
There is even potential for preventative health care using detection devices built with microfluidic technology. Devices used to detect particular pathogens are known as immunoassays. They make use of antibodies, which are proteins that bind to specific molecules, particularly those present on the surface of a pathogen. Microfluidic devices allow bound and unbound antibodies to be discriminated according to their different properties, indicating presence or absence of a pathogen. As with DNA sequencing, microfluidic devices are smaller, more portable, and faster at this specific task than a whole research lab full of equipment. It is claimed that immunoassays can detect E. coli in ground beef at a resolution of just one cell per gram. Perhaps in a few years’ time, these wallet-sized immunoassays will become an essential travel accessory, just like clean water tablets.
While microfluidic chips have been applied to solve many biological problems, its initial use in analytical chemistry has flourished and diversified. Now we have devices that are capable of advanced chemical synthesis and detection, even beyond the Earth.
The basic techniques and equipment in the chemist’s toolkit have remained largely unchanged since the first laboratory synthesis of urea in 1828, and in macroscopic situations miniaturisation of these processes is not necessary. However, one area where the importance of microfluidics is noticeable is in synthesizing nanomaterials.
The properties of nanoparticles are dependent on their size and shape. Current systems for growing nanoparticles show severe limitations, producing generally broad size distributions, leading to mixtures with unpredictable properties. The requirement for a specific particle size can supersede the need for large quantities, so growing nanoparticles more accurately, even at a small scale, is desirable.
Microfluidics is ideal for meeting this requirement. Not only does it allow rapid and controllable mixing, but variables such as temperature, concentration gradients and pressure can be manipulated to produce particles of specific size. Nanoparticles themselves have a wide range of applications: from drug delivery and medical diagnostics, to optical coatings and catalysis. The efficiency and accuracy that microfluidic devices lend to nanoparticle synthesis will allow us to make technological progress in diverse fields.
Chemical detection is another area where microfluidic principles can be applied, especially where the chemical of interest is present in a complex mixture at trace level. The primary focus for this type of technology is to optimise sensitivity and specificity of measurement in forms that are low cost, fully autonomous and miniaturised for portability. One of the main techniques targeted is that of gas chromatography (GC), a highly sensitive chemical analysis which is used to detect and quantify chemicals in air, water and soil.
A key part of existing GC systems is the fractionation column, which separates samples into their chemical components. These then pass through a detector to produce a chromatogram identifying the various chemicals. Typically the column is between 1.5 and 10 metres in length, making it far too bulky to transport.
For environmental testing, in particular atmospheric monitoring, samples are currently collected at remote locations and then returned to a laboratory for analysis. A small-scale, portable gas chromatography system would enable air quality to be analysed and recorded at the site of measurement, increasing the speed of response to any adverse changes. Microfluidics makes such a device possible as it enables miniaturisation of the GC column onto a single 10 centimetre square piece of glass.
Microfluidics has also allowed the miniaturisation of capillary electrophoresis, a separation technique that uses narrow-bore capillaries to separate molecules based on differences in charge, size and hydrophobicity. This is one of the proposed devices to be included on the European ExoMars rover mission scheduled for launch in 2013. The martian soil will be analysed for traces of biological compounds such as amino acids, the building blocks of proteins. The proposed experiment would separate amino acids from a soil sample, then use microfluidic capillaries to identify them by charge, size and crucially, chirality. This refers to the handedness of the amino acids which can exist in two mirror image forms, left-handed or right-handed. In simple chemical reactions, these molecules behave in the same way, but when it comes to complex biological reactions involving enzymes, the chirality matters. This is shown by the fact that all proteins on Earth are made up of left-handed amino acids. If the device on Mars finds an excess of amino acids of one particular chirality, it will be a clear sign they are biological in origin.
It would be impossible to send into space the amount of equipment required for this type of analysis in conventional setups, but with a microfluidic device, weighing only about two to three hundred grams, the search for life will be carried out right there on Mars.
A bit closer to home, devices based on the same technology are being used to police our own atmosphere. As many nations continue to be concerned about the threat of biological weapons, microfluidic methods are beginning to provide attractive antidotes to fear.
Small-scale detectors can now identify a broad range of chemical and biological agents. By protein fingerprinting, harmful bacteria and viruses are singled out. Biotoxins also feature in the vast catalogue of substances that new microfluidic devices are able to test for, including chemicals such as ricin and sarin which have both been associated with the military activities of the Cold War.
The USA has a strong focus on developing microfluidic technologies for defense applications. A number of American companies have invested and developed commercial biodefense solutions built on a microfluidic technology. The US Department of Homeland Security, as early as 2005, had already expressed interest in upgrading slow manual atmospheric testing facilities with automated microfluidic systems across major cities.
There seems to be no end to the new ways in which the use of microfluidic technologies can be integrated into our everyday activities. Earlier this year, researchers from Purdue University (Indiana, USA) discovered a new technique for conducting microfluidic analysis on paper. Using lasers to carve channels in the hydrophobic coating, it is hoped that this innovation will provide an even more inexpensive method to bring microfluidic technology to mainstream markets.
Microfluidics has established itself as an exciting field from which we can be sure that even more applications will emerge. As established technologies become ever more efficient and economical, the imaginative application of microfluidics to new technology will enrich and inform our understanding of our world and beyond.
Interview: Prof. Seth Fraden
Professor Seth Fraden is visiting from Brandeis University in Massachusetts. He specialises in soft condensed matter physics and is currently spending four months collaborating with members of the Department of Chemistry and Cavendish Laboratory in Cambridge. In America, his group has developed a new protein crystallisation technology using microfluidics, culminating in a device called the Phase Chip. He talks to BlueSci writer Vivek Thacker about his current work and future plans.
What started your interest in microfluidics?
SF: My background is in biological materials, looking at liquid crystals of viruses. Work on microfluidics began in my last sabbatical—at the time I was very impressed with the technological advances being made by Stephen Quake’s group at Caltech. He had developed a suite of microfluidic tools to synthesise small amounts of materials on a valve-based network, and he showed that it was a very scalable technology. I saw that as an advance to study my liquid systems in a very efficient manner, and decided to pick this up in my upcoming sabbatical.
Is it easy to scale up from a microfluidic system to a bulk system?
SF: No! Because the physics is different, you do not want to have the intention of scaling up. But the technology is scalable. Microfluidic valves are made photo-lithographically, like printing, so the effort to make a hundred is the same. It is like semiconductor manufacturing—once you’ve learnt to make one transistor, you can make ten million of them on the same wafer.
Your Phase Chip carries out protein crystallisation. Is this something some labs focus on particularly?
SF: Some labs do focus on making protein crystals, but their interest is not on the crystallisation process itself. They just want to use the crystals in diffraction experiments and obtain the structure of the proteins.
If I am going to make a contribution to the field, the product has to leave my lab. If the chip costs $1000, then it will not leave my lab because it is not commercially viable. Since realising this, I have focussed on the question: can we make a device which retains the essential qualities but is 100 times cheaper?
How do you propose to do that?
SF: You can achieve protein crystallisation with discrete components. Before we had one integrated device that did everything, but now you can make one optimised device for each step. To really tackle the protein crystallisation problem, since the crystals we produce are so small, we have to develop a whole new suite of technologies to complement ours, so I have set up a collaboration with a synchrotron beam scientist who specialises in diffraction experiments. The whole community is converging on this idea because it is clear that small crystals are easier to make than bigger ones.
Where do you see microfluidic technology going in the future?
SF: Microfluidics will have applications in a large set of devices, but they will definitely be under-the-hood as components. The first ten years of the field were focussed on building extended microfluidic devices, but the future is going to be integration into larger systems.
Helen Gaffney is a second year Natural Sciences Tripos student
Wendy Mak is a PhD student in the Department of Physics
Lindsey Nield is a PhD student in the Department of Physics
Vivek Thacker is a PhD student in the Department of Physics