SUNDAY, 19 MARCH 2017
In 1947, in a post-war world divided by intense political tensions, scientists from countries all over the planet joined their effort to tackle together one of the most serious threats to human health: influenza.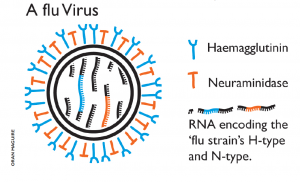 Image by Oran Maguire. Influenza viruses are RNA viruses that make up three of the five genera of the family Orthomyxoviridae. Type A influenza viruses are the most virulent among the different influenza types and cause the severest disease. Wild aquatic birds are their natural hosts.
Image by Oran Maguire. Influenza viruses are RNA viruses that make up three of the five genera of the family Orthomyxoviridae. Type A influenza viruses are the most virulent among the different influenza types and cause the severest disease. Wild aquatic birds are their natural hosts.Today, the production of an annual flu vaccine that keeps pace with an evolving and sophisticated virus, preventing thousands of deaths each year, poses an enormous challenge to science.
Each winter, a significant proportion of individuals are vaccinated or infected with flu. They develop immunity through the production of antibodies that bind to the haemagglutinin (HA) protein on the surface of the virus. Individuals retain the capacity to produce these specific antibodies for many years. Thus, in theory, the next winter there should be fewer individuals who are susceptible to infection – but alas, this is not the case. Between winters, the virus HA gene mutates, giving rise to amino acid changes that alter aspects of the HA molecule structure. The next winter, the virus is once again unrecognisable to the immune system: a new flu outbreak emerges. Science must monitor the changing antigenicity of the virus and aim to accurately predict the likely amino acid changes that will be seen in the virus the following year.
To add to the complexity, there is not just one, but four strains of influenza virus currently circulating significant proportions: the Influenza A H3N2 and H1N1 strains and the Influenza B Yamagata and Victoria strains. Almost a year ahead of time, scientists must accurately predict the future antigenic behaviour of the virus, and it has to be right. This is a challenge that requires extensive collaboration from laboratories all over the world to be solved.
In essence, the process of influenza surveillance involves monitoring influenza samples globally, analysing their DNA sequences and antigenicity and pooling this global data to decide the three virus strains (one H3N2, one H1N1 and one influenza B) that will be propagated and packaged into vaccines.
At the front line, doctors’ surgeries around the world collect samples from patients with flu-like symptoms. In fact, Trumpington Street Medical Practice is the assigned surgery for Cambridge. These surgeries send their samples to National Influenza Centres, where scientists perform preliminary analyses of the influenza samples. They then send a representative selection of these to one of six Global Influenza Collaboration Centres.
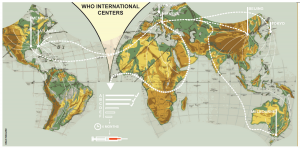 Image by Oran Maguire
Image by Oran Maguire“Alone we can do so little; together we can do so much.” - Helen Keller
As well as sequencing data, gathering information on the antigenicity and structure of the HA molecule is crucial. A set of standard protocols is followed at the six Global Influenza Collaboration Centres to determine the extent to which the influenza viruses have changed antigenically since the previous winter. Practically, this involves using post-immunisation ferret antisera (serum from ferrets that have been infected with influenza, containing anti-flu antibodies) in a procedure known as a haemagglutination inhibition assay to reveal how well the antibodies in the serum can recognise the new viruses.
These data, from thousands of viruses collected around the world, are compiled and sent to the World Health Organisation (WHO) headquarters in Geneva. In February and September of each year (for the Northern and Southern Hemisphere vaccines respectively) global influenza experts meet for three days to comb through the data and vote on the three vaccine candidates they believe will be the most effective vaccine strains. Get it right, and the vaccine offers 60% effectiveness. Get it wrong, and the vaccine has very little to no efficiency – this was for example the case during the winter of 2014/2015. Yet the success of global influenza surveillance as a feat for science, globalization and medicine cannot be ignored.
The influenza surveillance program is a compelling example of what science, and the world, can achieve when truly globalised
Strikingly, global influenza surveillance began in 1947 just after the end of World War II. Scientists at the National Institute of Medical Research in Mill Hill, London, realised they needed to begin monitoring the virus in order to make effective vaccines. Christopher Andrewes, one of the scientists who first isolated the influenza virus, set up a network of laboratories for the WHO, with the Worldwide Influenza Centre at Mill Hill at the centre of the project. Since then, the surveillance process has evolved to the highly sophisticated, global network we now know as GISRS (Global Influenza Surveillance and Response System).
 Image by Oran Maguire
Image by Oran MaguireThe success of the influenza vaccine and the cooperative efforts that go into its production serve as a reminder that great things can be achieved when we work together. The establishment of collaborative structures and distribution of resources provides an efficient network in which an effective influenza vaccine is produced every six months. There are important lessons to be learned here for managing similar such global outbreaks, including Zika, Ebola and HIV.
Beyond science, in a world where globalisation is being rejected by electorates worldwide and nationalism is the word of the day, global influenza surveillance assumes a new significance. In the coming years, in an increasingly fractured and defiant political landscape, it may be the duty of science to lead the way in promoting international collaboration and cooperation.
