MONDAY, 25 NOVEMBER 2013
The brain is a complex structure comprised of thousands of neurons and glia (cells that support and protect nerves). Neurons come in many shapes and sizes and have a number of different purposes. In order for the brain to function properly each neuronal type must be generated at the correct time during development, in a precise number and in the appropriate brain region. This process facilitates the right connections with other neurons towards formation of sophisticated neuronal circuits. Understanding how the brain achieves this intricacy with such accuracy remains one of the major challenges in the field of neuroscience.
The overwhelming complexity of the brain makes it a difficult organ to study. However, the eye retina, an outpocket of the forebrain, has proven to be a powerful—and most importantly accessible—model for neuronal development studies. The retina has a relatively simple structure, consisting of five neuronal cell types (photoreceptors, horizontal cells, bipolar cells, amacrine cells and retinal ganglion cells), which are responsible for processing and transmitting visual signals to the brain via the optic nerve. Within the eye, neurons are highly organised into three separate layers. We know this precise architecture is crucial for proper function, however, little is known about how it is produced. The retina is formed during embryonic development from progenitor cells. These cells express genes which allow them to follow one of a number of different neuronal fates and ultimately differentiate into mature neurons. However, exactly how a cell’s fate is decided is not known.
The use of microscopy has revolutionised our understanding of the developing nervous system. Cells can be visualised by labelling them with dyes, or more commonly, adding genes that code for fluorescent proteins, which are produced only when the cells coexpress a certain gene. For example, if the expression of a green fluorescent protein is driven by the expression of PTF1A, a gene expressed only in amacrine and horizontal retina neurons, only these two cell types will be labelled green. Although traditional labelling techniques have enabled high resolution 3D imaging of entire vertebrate brains, these methods can only be performed in non-living tissues, meaning that they only capture a snapshot of development. Observing distinct cell populations in real time as they go through the developmental process has remained a challenge.
These difficulties can be overcome using the embryos of the zebrafish, a small tropical freshwater fish, very popular in scientific research. Zebrafish embryos are transparent, allowing brain development and neuronal activity to be studied in real time in the whole live organism using imaging microscopy.
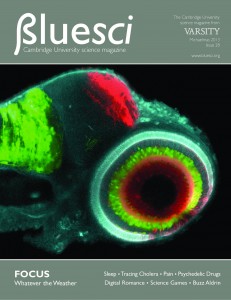
The image on the cover of BlueSci Issue 28 shows a four day-old zebrafish embryo that has been genetically modified to allow each cell type within the retina to be identified by a uniquely coloured fluorescent protein. This tool, termed the SoFa fish (spectrum of fates approach), has been generated by Dr Xana Almeida in Professor Bill Harris’ lab in the Department of Physiology, Development and Neuroscience at the University of Cambridge, and it allows retinal development to be visualised in real time in the living animal. Photoreceptors and bipolar cells are labelled in cyan, amacrine and horizontal cells are green and retinal ganglion cells are red.
What distinguishes the SoFa fish from other labelling techniques is that the expression of the fluorescent markers in the retinal cells is switched on by genes that become active as progenitor cells commit to differentiation. This means that we can visualise what a cell will become before it has fully differentiated. By studying the SoFa fish retina with high-resolution confocal microscopy, the researchers witnessed for the first time the birth of all five types of retinal neurons in live zebrafish embryos, as it was happening.
This ability to image the developing retina in 4D (with time being the 4th dimension) provides an unprecedented view of neural development, and will open the door to many further studies. For example, researchers can now determine patterns of cell division and migration, allowing them to study how the architecture of the retina is created. Additional applications are visualising the establishment of connections between neurons, as well as monitoring the effects of altered gene expression on cell fate. Therefore, the SoFa fish retina represents an invaluable tool to understand the complex sequence of events required to create functional tissues.
Dr Xana Almeida’s image was one of three winning entries in the University of Cambridge Graduate School of Life Sciences (GSLS) image competition. The competition, run during the Cambridge Science Festival, showcases the variety of biological research in Cambridge. To fi nd out more, visit the GSLS website. The other winning images, taken by Nuri Purswani and Mubeen Goolam, can be found on www.bluesci.org.